Biofilms are ubiquitous surface-attached bacterial communities embedded in an extracellular matrix. Biofilms can cause major problems: in medicine, biofilms underlie chronic infections that are difficult to eradicate; in industry, biofilms foul surfaces of pipes and clog filtration devices. By contrast, biofilms can be beneficial, for example, as a normal part of the human microbiota and in wastewater treatment. Despite extensive work to identify biofilm components and to discover how they are regulated, we have little understanding of how cells are arranged within a biofilm and how the matrix components interact to form these unique living materials. To fill these gaps in our knowledge, we are developing new experimental and theoretical tools to generate a comprehensive understanding of bacterial biofilms from the single-cell level to the community level. Such an understanding is crucial to combat biofilm-related problems and to harness beneficial biofilms. Below are some specific questions we are pursuing using Vibrio cholerae as the model biofilm forming species. Click links below for more information.
I. Cellular-scale architecture of biofilms
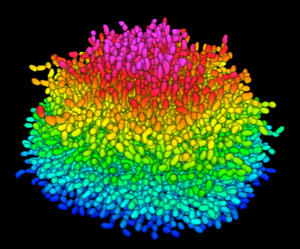 |
We aim at understanding the biofilm formation process at the single cell level. We are developing new imaging and computational tools to investigate how bacterial cells construct a 3D biofilm cell by cell. Click here for more information. |
II. Mechanomorphogenesis of biofilms
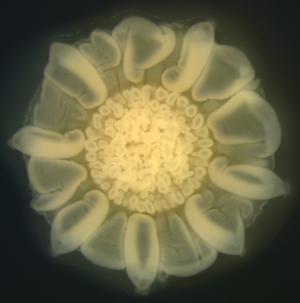 |
We investigate how mechanical forces shape the morphological development of biofilms growing on various surfaces. Click here for more information. |
III. Biofilms as living, active materials
 |
We are interested in how biofilm material properties arise from individual components, and how growing biofilms differ from classic, inanimate materials. Click here for more information. |
IV. Ecology and evolution of bacterial biofilms
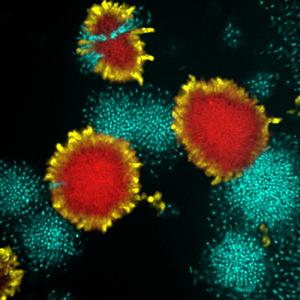 |
We are generally interested in what intrinsic genetic and mechanical mechanisms and what external factors control the social ecology inside these microbial communities. Click here for more information. |